
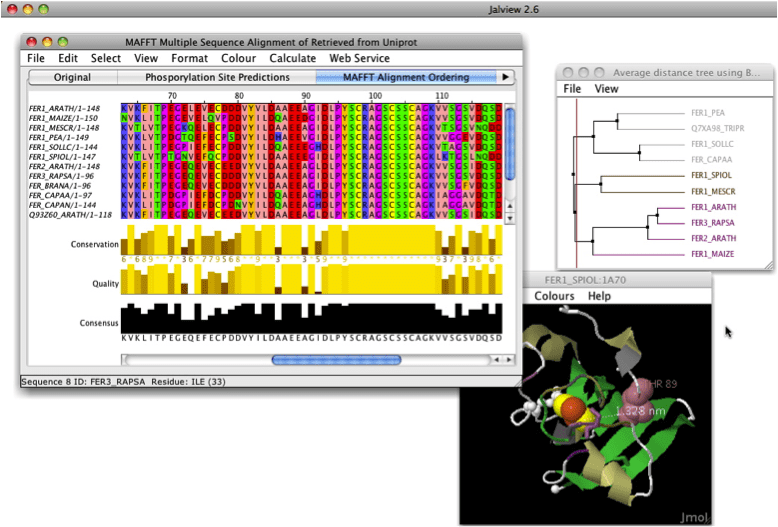
The second step uses this tree to guide a progressive alignment. The first step constructs a distance matrix between pairs of sequences using k-mer clustering, this is then converted into a tree. MUSCLE is said to have four major steps in its alignment process. Boasting both speed and accuracy, it compares very favorably to other multiple-sequence alignment programs. It joins Clustal, making it the second MSA program in Sequencher’s DNA-Seq Tools. MUSCLE, a multiple-sequence alignment (MSA) program, joins the Sequencher 5.1 family of plugins. Speed up your Clustal alignments by combining Clustal with the power of Sequencher's Assemble by Name functionality for aligning multiple sequences from different sources.įinally, export the results in a variety of different formats such as MSF, Phylip, NEXUS and FastA for use in other programs or simply create a consensus and export it. Once the alignment is complete you will see the results as a contig or contigs within Sequencher which you can subject to a variety of analyses. Choose from a range of parameters to control the alignment process. You can use Clustal to align your sequences directly from the Sequencher project window. It is a widely used multiple-sequence alignment program which works by determining all pairwise alignments on a set of sequences, then constructs a dendrogram grouping the sequences by approximate similarity and then finally performs the alignment using the dendogram as a guide. 27(14):2001-2002).Clustal has been part of the Sequencher family of plugins since version 4.9. JABA Web Services can be accessed from the Jalview desktop application and providemultiple alignment and sequence analysis calculations limited only by your own local computing resources. A new multiple sequence alignment service forClustal Omega is also provided, in addition to standard JABAWS:MSA services for Clustal W, MAFFT, MUSCLE,TCOFFEE and PROBCONS. JABAWS v2.0introduces protein disorder prediction services based on DisEMBL, IUPred, Ronn, GlobPlot and proteinsequence alignment conservation measures calculated by AACon. JABAWS 2 - provides web services for multiple sequence alignment, prediction of protein disorder, and aminoacid conservation conveniently packaged to run on your local computer, server or cluster. This program has proved very useful in recent studies on the classification of bacterial viruses. The program will provide Bidirectional Best Hit, OrthoMCL or COGTriangle results.
PairAlign (EzBioCloud, Seoul National University, Republic of Korea)- pair wise nucleotide sequence alignment for nucleotide sequences 20 accession numbers can be assessed. VerAlign multiple sequence alignment comparison is a comparison program that assesses the quality of a test alignment against a reference version of the same alignments. Wasabi - (Andres Veidenberg, University of Helsinki, Finland) is a browser-based application for the visualisation and analysis of multiple alignment molecular sequence data. LAST - provides a lot of control of data handling, along with dotplots and coloured alignments ( Reference: Kielbasa SM et al. SFESA ( Shift to Fix secondary structure Element S in Alignments) - is a web server for pairwise alignment refinement by secondary structure shifts.SFESA evaluates alignment variants generated by local shifts and selects the best-scoring alignment variant. LALIGN shows the alignments and similarity scores, while PLALIGN presents a "dot-plot" like graph. 39(Web Server issue):W38-44)Ĭompare Two Sequences with LALIGN/PLALIGN find internal duplications by calculating non-intersecting local alignments of protein or DNA sequences.
#Software to align dna sequences series
The user can, through a series of tabs, navigate multiple results pages, and also includes novel functionality, such as a dotplot graph viewer, modeling tools, an improved 3D alignment viewer and links to the database of structural similarities.
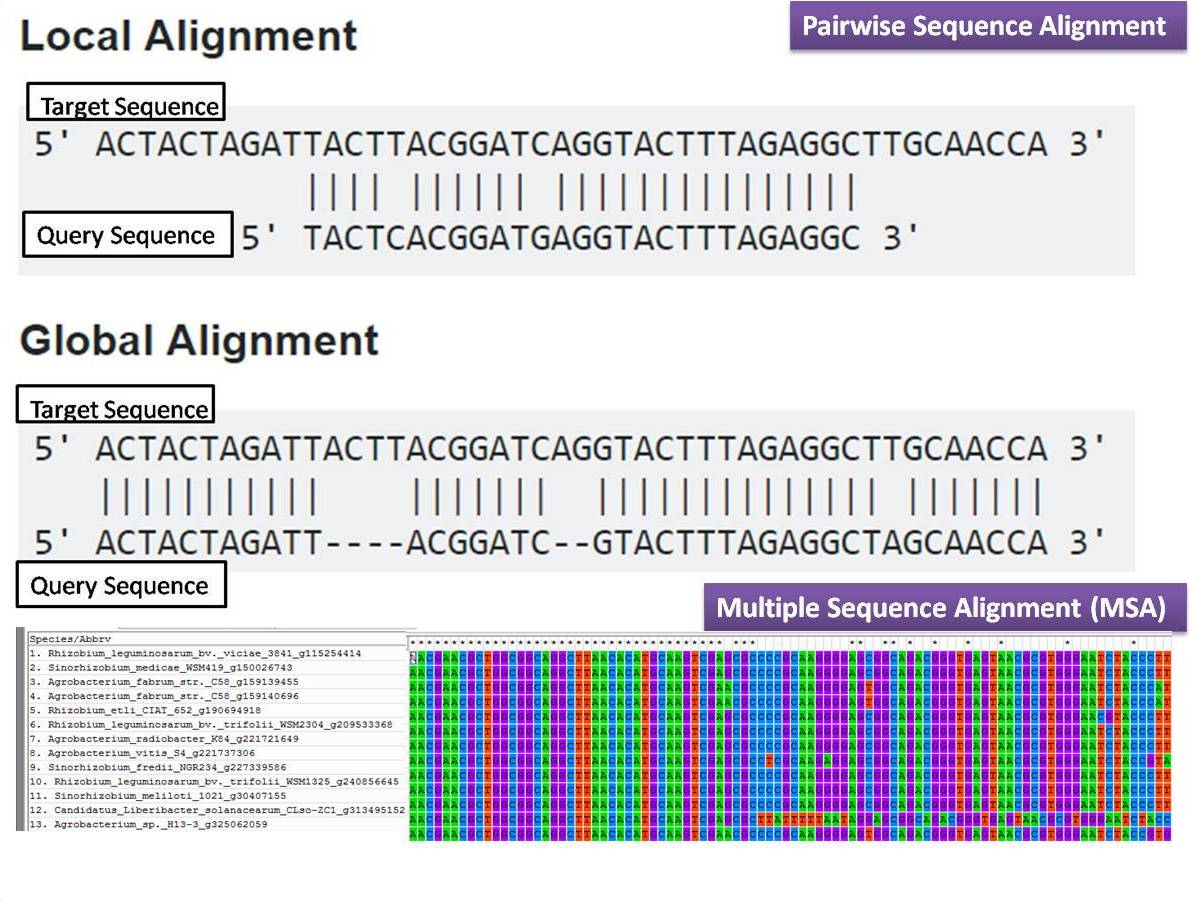
The profile of a user's protein can now be compared with ~20 additional profile databases. Provides one with % identity for different subsegments of the sequence.įFAS - The Fold and Function Assignment System. LALIGN - (EMBL-EBI) finds multiple matching subsegments in two sequences. Alignments ALIGNMENTS COMPARE TWO SEQUENCES :


 0 kommentar(er)
0 kommentar(er)
